We have a new paper out in Molecular Biology and Evolution. See the press release here.
When the honey bee Apis mellifera genome was sequenced in 2006, researchers found that the bee has fewer innate immune genes – the genes that recognize and destroy pathogens – than other solitary insects. The bee had about 30% of the innate immune genes found in flies and mosquitos. Wow! Why?
Dr. Jay Evans (USDA) and colleagues hypothesized that maybe bees have fewer pathogens, or that they can fight pathogens better with social behaviour; worker bees can groom themselves and their sisters, and they can recognize sick/dead larva and remove them from the colony.
Brock Harpur, a MSc (now PhD) student in the lab is very interested in immunity in social insects, and he decided to study the evolution of 13 innate immune genes in the honey bee. We sequenced these genes in about 40 different honey bee workers, and in one worker of the closely related Asian honey bee Apis cerana. This allowed us to discover mutations in innate immune genes within the European honey bee Apis mellifera, and between the European and the Asian honey bee (Apis mellifera vs. Apis cerana). We compared the amount of mutations in innate immune genes to those found in 20 randomly chosen genes.
The DNA sequence of a given gene codes for a specific sequence of amino acids that get folded into a 3-dimensional protein that then preforms a specific function within the cell. Mutations in a gene sequence come in two flavours: there are mutations that don’t change the amino acid sequence of the resulting protein (we call these silent mutations), and there are mutations that change the amino acid sequence of the resulting protein (we call these replacement mutations). The silent mutations – because they do not affect the shape and structure of the resulting protein – do not often affect fitness and are not ‘weeded-out’ by natural selection; silent mutations accumulated within species, and between species. However, replacement mutations will change the shape / structure of proteins, and often the resulting protein will not function optimally (if it works at all). These mutations, because they are often bad, are then rapidly removed by natural selection (because bees with these mutations do not survival as well).
Now imagine a gene sequence that was functional in an ancestor but is now no longer necessary. Replacement mutations in such genes will no longer be weeded-out by natural selection (because bees with these mutations survival normally), and these mutations can then accumulate similar to silent mutations; this process is called ‘relaxation of purifying selection or relaxation of constraint’.
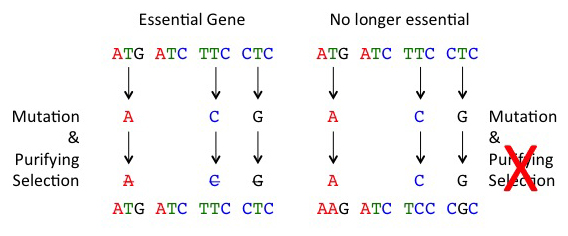
When selection is relaxed (right), mutations accumulate at fast rates in gene sequences. Normally, mutations that alter the amino acid sequence of a protein will be removed by purifying selection as a large fraction of these mutations are bad (left)
This is exactly what we see when we look at most immune genes in the honey bee. When we examine silent mutations, immune genes and random genes have similar rates. However, if we examine replacement mutations, we find that immune genes have 3 to 4 higher mutations relative to random genes; this is consistent with a relaxation of purifying selection. We reckon that the same process that resulted in massive loss of innate immune genes in the bee is still acting to erode some of the remaining innate immune genes in contemporary bee populations. Our work supports Dr. Jay Evans’ idea that some aspect of social living makes an individual-based innate immune system less useful.